Current PhD vacancies
Next call starts on "Mon, 16.Mar 2026"

- Cancer
- Cardiovascular systems
- Endocrinology and Metabolism
- Molecular and cell biology
Cachexia is a multifactorial syndrome characterized by severe muscle wasting that accompanies various diseases, including cancer. It represents a major prognostic factor and a leading cause of mortality among cancer patients. The severity of cachexia correlates with circulating levels of the cytokine interleukin-6 (IL-6); however, the mechanisms driving skeletal and cardiac muscle atrophy remain poorly understood.
We hypothesize that cardiac cancer cachexia arises from direct actions of tumour-derived IL-6 and IL-6-like cytokines on cardiac myocytes. Specifically, we propose that cytokine-induced signalling enhances sarcoplasmic reticulum SERCA activity, leading to elevated ATP turnover and energy expenditure. This increased energetic demand may underlie the elevated metabolic rate and catabolic processes responsible for cardiac atrophy.
To test this, we will combine electrophysiology and confocal imaging to assess cardiac myocyte Ca²⁺ handling, quantify ATP levels and metabolic rates using Seahorse assays, and evaluate cardiac performance through isolated heart preparations, echocardiography, and PET/CT imaging. Molecular studies will include IL-6 knockdown or knockout (shRNA/CRISPR-Cas9), cytokine profiling, cardiac-specific STAT3 deletion, RNA-seq, ATAC-seq, and western blotting to trace downstream IL-6 signalling. Findings will be validated in a melanoma model of cancer cachexia and in samples from human cancer patients.
This study introduces a novel concept that cardiac cachexia originates from cytokine-mediated increases in energy demand, providing new mechanistic insight and potential therapeutic avenues against cancer-associated cachexia.
What you’ll work on
- Explore how cytokine-driven calcium handling leads to cardiac atrophy in cancer cachexia.
- Combine electrophysiology, imaging, and molecular methods to study heart metabolism and function.
- Work hands-on with mouse models and advanced imaging techniques.
- Collaborate with researchers in molecular signal transduction and translational medicine.
What you’ll learn (and get really good at)
- Electrophysiology and confocal calcium imaging.
- Animal experimentation and functional heart analysis (echocardiography, PET/CT, working heart).
- Molecular profiling (RNA-seq, ATAC-seq, western blotting).
- Experimental design, data analysis, and reproducible scientific workflows.
Your research environment
You’ll be part of the Molecular Signal Transduction network and the Young Scientist Association (YSA), giving you access to interdisciplinary collaborations, training, and mentoring in an international research community.
Master’s degree (or equivalent) in physiology, molecular biology, biomedical sciences, or a related field.
Advantage: Experience in animal models, electrophysiology, or confocal calcium imaging.

- AI/ML
- Biomedical engineering
- Computer lab
- Human research
AutoPiX connects clinical expertise, multimodal imaging and cutting-edge AI to address urgent unmet needs and provide more precise an personalized care in arthritis – a group of chronic diseases that cause inflammation, joint destruction and disability. The project unites one of the world’s largest curated arthritis imaging collections (>100,000 X-ray, ultrasound and MRI scans across sites and longitudinal timepoints) with rich clinical and laboratory metadata, and an international IHI-funded consortium of academia, pharma, medtech, startups and patient partners.
Your Mission as PhD is to develop novel machine-learning methods to (1) detect small, clinically meaningful changes in joints, (2) enable robust longitudinal monitoring and treatment-response prediction, and (3) build a multimodal Visual Foundation Model for arthritis that boosts downstream tasks and enables discovery of new biomarkers and arthritis subtypes. Ultimately, this should lead to earlier diagnosis, improved monitoring and better-tailored treatments. Technical directions include self-supervised and multimodal representation learning, explainability and domain-robustness across sites and devices.
This role is ideal for a candidate who is passioned about self-driven high-impact ML research with immediate clinical translation, access to large, heterogeneous real-world data, strong compute resources, industry collaborations and mentorship aimed at publishing high-impact ML/medical-imaging papers and clinical impact.
What you’ll work on
- Develop and benchmark novel machine learning models for detecting subtle, clinically relevant changes in arthritis imaging.
- Create robust algorithms for longitudinal disease monitoring and treatment-response prediction.
- Build a multimodal Visual Foundation Model for arthritis that integrates imaging, clinical and laboratory data.
- Advance methods for explainability, domain robustness and federated or privacy-preserving analysis.
- Collaborate with clinicians, imaging experts, data scientists and industry partners to ensure clinical translation.
- Present and publish your research at top-tier ML and medical imaging conferences.
What you’ll learn (hands on skills)
- Advanced ML research skills: multimodal representation learning, foundation model design, explainability and domain adaptation.
- Handling and analyzing large, multi-site medical imaging datasets across modalities (X-ray, ultrasound, MRI).
- Scalable ML workflows: GPU-based training, experiment tracking, reproducible pipelines, model validation and deployment.
- Research excellence: hypothesis formulation, experimental design, statistical evaluation, scientific writing and presentation.
- Translating clinical problems into AI tasks and working effectively across disciplines with clinicians, industry and patient partners.
Your research environment
You will be embedded in the Computational Imaging Research Lab (CIR), affiliated with the Comprehensive Center for AI in Medicine (CAIM) and the Division of Rheumatology at the Medical University of Vienna, and will be an integral part of the international AutoPiX consortium (academia, pharma, medtech, startups, patient partners).
Key benefits:
- Close collaboration with other AutoPiX PhD students, regular joint training, workshops, and research retreats.
- Great Work Environment: Friendly, international team in one of the world’s most livable cities, with health programs, an on-site cafeteria and excellent public transport.
- Top-Tier Mentorship: leading experts/supervisors in AI, rheumatology and clinical imaging, for high-impact publishing.
- Exceptional resources: access to one of the largest curated arthritis imaging datasets and high-performance GPU clusters.
- Career & real-world impact: access to conferences, strong pathways into academia, industry R&D or clinical-translational roles, plus networking with patient advocates to ensure clinical relevance.
- Master degree (MSc or equivalent) in Computer Science, Machine Learning or a related discipline.
- Strong foundation in machine learning / deep learning (theory + hands-on). Evidence may include a thesis, open-source project, or publications.
- Programming/engineering expertise. Proficient in Python and practical with a deep-learning framework (PyTorch or TensorFlow). Comfortable writing clean, reproducible code and using version control.
- Analytical mindset & problem-solving drive.
- You are passionate about formulating hard research questions, designing experiments, and iterating to robust solutions.
- You enjoy interdisciplinary collaboration, mentoring and sharing knowledge with peers and students.
Advantage:
- Prior experience with medical imaging or multi-site clinical datasets.
- Experience with multimodal or self-supervised learning, longitudinal modelling, foundation-models, or explainable AI.
- Familiarity with training on GPU clusters, experiment tracking, containerization, and reproducible pipelines.
- Evidence of research potential for top venues (e.g., first-author papers, strong project results).
- German is a bonus

- AI/ML
- Bioinformatics
- Computer lab
AutoPiX connects clinical expertise, multimodal imaging and cutting-edge AI to address urgent unmet needs and provide more precise a personalized care in arthritis – a group of chronic diseases that cause inflammation, joint destruction and disability. The project unites one of the world’s largest curated arthritis imaging collections (>100,000 X-ray, ultrasound and MRI scans across sites and longitudinal timepoints) with rich clinical and laboratory metadata, and an international IHI-funded consortium of academia, pharma, medtech, startups and patient partners. Your role as a PhD candidate is to develop novel machine-learning methods to (1) detect small, clinically meaningful changes in joints and related tissues, and (2) identify new imaging biomarkers suitable as clinical endpoints. You will have the freedom to explore state-of-the-art approaches such as self-supervised pretraining, foundation models, domain adaptation, image registration, uncertainty modeling, and multimodal fusion with patient data. Beyond algorithm development, you will contribute to a data-centric workflow, helping to decide which images to annotate, refining data quality control, and continuously improving the dataset through iterative model feedback. The goal is to deliver models that are clinically validated in trials and ready for deployment in patient care. This role is ideal for a candidate who is passionate about self-driven high-impact ML research with immediate clinical translation, access to large, heterogeneous real-world data, strong computeational resources, industry collaborations and mentorship focused on publishing high-impact ML/medical-imaging papers as well as achieve concrete clinical impact.
What you’ll work on
- Develop and evaluate machine-learning methods to detect small, clinically meaningful changes in joints and related tissues on X-ray, ultrasound, and MRI.
- Design models for longitudinal disease monitoring and treatment-response prediction, including image registration and temporal modeling.
- Build multimodal models—up to a visual foundation model—that integrate imaging with clinical and laboratory data to derive candidate imaging biomarkers suitable as clinical endpoints.
- Apply and assess approaches such as self-supervised pretraining, domain adaptation, uncertainty estimation, and multimodal fusion; document assumptions and limitations.
- Contribute to a data-centric workflow: select images for annotation, refine data quality control, and improve the dataset iteratively using model feedback.
What you’ll learn (hands on skills)
- Understanding of rheumatic diseases: linking imaging findings with underlying disease mechanisms, inflammation patterns, and treatment response.
- Advanced ML research skills, including developing novel ML methodology Experience in working with large medical imaging datasets and developing models for multi-site, multi-device medical data. Practical skills: scalable training on GPU clusters, experiment tracking, reproducible pipelines, model deployment and validation.
- Research competencies: experimental design, statistical validation, scientific writing, and presenting at international conferences.
- Interdisciplinary collaboration: translating clinical questions into ML tasks, working with clinicians, industry partners and patient partners.
Your research enviroment
You will be part of a newly established AI Research group at Division of Rheumatology, Department of Medicine III, Medical University of Vienna (Prof. Peter Mandl), and embedded in the Comprehensive Center for Artificial Intelligence in Medicine (Prof. Georg Langs), who will also serve as your senior supervisor. You will be an integral part of the international AutoPiX consortium (academia, pharma, medtech, SMEs, patient research partners and regulatory agencies).
Key benefits:
- Close collaboration with other AutoPiX PhD students, regular joint training, workshops, and research retreats.
- Great Work Environment: Friendly, international team in one of the world’s most livable cities, with health programs, an on-site cafeteria and excellent public transport.
- Top-Tier Mentorship: leading experts/supervisors in AI, rheumatology and clinical imaging, for high-impact publishing.
- Exceptional resources: access to one of the largest curated arthritis imaging datasets and high-performance GPU clusters.
- Career & real-world impact: access to conferences, strong pathways into academia, industry R&D or clinical-translational roles, plus networking with patient advocates to ensure clinical relevance.
• Master’s degree (MSc or equivalent) in Computer Science, Mathematics, Machine Learning or a related discipline.
• Strong foundation in machine learning / deep learning (theory + hands-on). Evidence may include a thesis, open-source project, or publications.
• Programming/engineering expertise. Proficient in Python and practical with a deep-learning framework (PyTorch or TensorFlow). Comfortable writing clean, reproducible code and using version control.
• Analytical mindset & problem-solving drive. You are passionate about formulating hard research questions, designing experiments, and iterating to robust solutions.
• Teamwork and Communication. You enjoy interdisciplinary collaboration, mentoring and sharing knowledge with peers and students. You communicate clearly in spoken and written English. German is not required but is considered a bonus.
Advantage:
• Prior experience with medical imaging and/or multi-site clinical datasets.
• Experience with multimodal or self-supervised learning, longitudinal modelling, foundation-models, or explainable AI.
• Familiarity with training on GPU clusters, experiment tracking, containerization, and reproducible pipelines.
• Evidence of research potential for top venues (e.g., first-author papers, strong project results).

- Animal research
- Biochemistry
- Molecular and cell biology
- Wet lab
Limb-girdle muscular dystrophies (LGMD) comprise a group of rare, genetically heterogeneous neuromuscular disorders characterized by progressive weakness of the shoulder and pelvic muscles. Although next-generation sequencing has advanced genetic diagnosis, many patients still lack a definitive molecular explanation. A recent nationwide study in Austria identified several cases with Variants of Uncertain Significance (VUS)—genetic alterations whose functional impact remains unclear. These variants affect proteins involved in skeletal muscle or motor neuron function, making them strong candidates for further investigation.
Two PhD projects will explore the functional consequences of selected VUS using an integrated, multi-level experimental strategy that combines molecular analyses, histopathology, iPSC-derived muscle models, and in vivo mouse studies to closely replicate human disease mechanisms. The findings are anticipated to advance patient counseling and therapeutic development, while also establishing a model framework for research into rare neuromuscular diseases more generally.
What you’ll work on
- Functionally characterize VUS linked to LGMD using molecular analyses, histopathology, and iPSC-derived muscle models
- Translate findings in vivo using relevant mouse models to mirror human disease mechanisms
- Integrate bioinformatics with experimental readouts to pinpoint pathogenic mechanisms
- Present results, co-author manuscripts, and help shape future study directions
What you’ll learn (and get really good at)
- iPSC workflows: culture, differentiation, and assay design for skeletal muscle models.
- Muscle cell biology: setup and execution of functional muscle analyses.
- Bioinformatics: variant interpretation and multi-level data integration.
- In vivo experiments: design, conduct, and analysis in relevant mouse models.
- Scientific exchange: work within a multidisciplinary, collaborative setting and engage with an international network, incl. the MedUni interest group “The Neuromuscular System.”
Depending on your interests/experience, you can lean more into the wet-lab/iPSC track or the computational/bioinformatics track—while gaining fluency across both.
Your research network
You’ll join a collaborative, interdisciplinary team led by Lilli Winter and Ruth Herbst, working closely with Inga Konezcny and Hakan Cetin, whose expertise spans neuromuscular disease and translational research. You’ll be enrolled in the Molecular Signal Transduction PhD program, gaining connections to related groups across the field, and you’ll engage with the MedUni interest group “The Neuromuscular System” for seminars, exchanges, and community-building around muscle biology.
• Master in Cell Biology, Biochemistry, Molecular Biology or equivalent
• Advantage: Experience in protein biochemistry, cell culture, bioinformatics, microscopy, and mouse work; an animal work certification (FELASA or equivalents) is also necessary, but can be obtained in the initial phase of the project.
• Interest in evaluating skeletal muscle diseases

- AI/ML
- Cardiovascular systems
- Epidemiology
- Mathematics and/or Statistics
- Public health
Chest pain is one of the most common symptoms in the emergency medical setting. While certain differential diagnoses, such as acute coronary syndrome, acute aortic syndrome, or pulmonary embolism, are potentially life-threatening, the majority of cases are attributable to non-critical causes, such as musculoskeletal origin. Supporting fast and well-informed decisions, making sure critically ill patients are treated at the right place in a timely manner, as well as identifying those not at acute risk, requires a sound foundation in data and models. Together we will analyze and develop clinical pathways for those patients, and further diagnostic and prognostic models. Our research will primarily draw upon observational data from international collaborative networks.
What you’ll work on
- Turn real-world healthcare data into insights that improve triage and care pathways for chest-pain patients.
- Build and evaluate diagnostic and prognostic models to support rapid, well-informed decisions in emergency settings.
- Collaborate with clinicians, data scientists, and international partners to translate methods into practice.
- Share your work at conferences and through publications.
What you’ll learn (and get really good at)
- Working with routine healthcare data and international observational datasets (data curation, linkage, quality, documentation).
- Depending on your interests/experience, either
- federated data analysis and privacy-preserving modeling, or
- methodological advances in diagnostic/prognostic research (model design, validation, calibration, transportability).
- Reproducible research workflows (coding best practices, version control, transparent reporting).
Your research environment
We are part of the university’s Comprehensive Center for AI in Medicine and serve as the national node of the EMERGE research network. All projects are conducted with international collaborators, and you’ll regularly participate in conferences across Europe.
Required: Master’s degree (or equivalent) in a relevant field (e.g. medical informatics, medical statistics, or public health)
Advantage: Experience with one or more of the following: health services research, research with observational or routine health data, statistical methods of diagnostic or prognostic research
- Bioinformatics
- Biomedical engineering
- Computer lab
- Mathematics and/or Statistics
- Medical physics and/or Biophysics
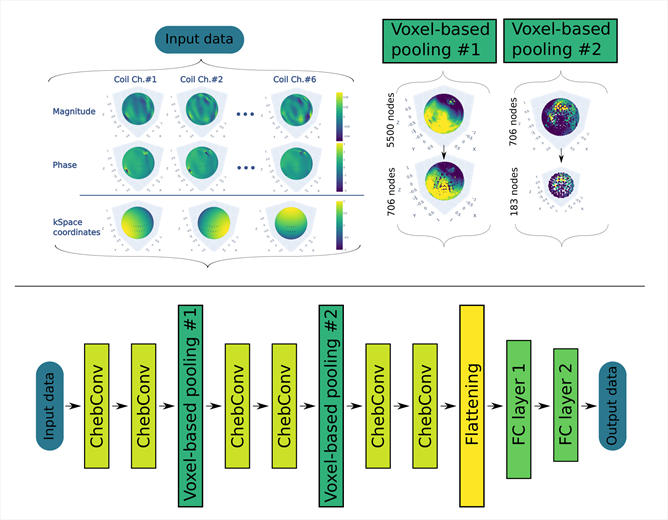
This FWF-funded PhD project aims to develop a deep learning-driven method for real-time motion correction in MRI. The candidate will design and implement ultra-fast (<10ms) k-space MR navigators to estimate 6 DoF - head motion via Deep Learning. The project will combine MR physics, signal processing, and AI to enable high-quality imaging free from motion artefacts at ultra-high field MRI systems.
What you’ll work on
- Develop and benchmark deep learning models for real-time head motion detection from ultra-fast k-space MR navigators
- Design and optimize 3D MR navigator trajectories for accurate six-degree-of-freedom (6-DoF) motion estimation
- Integrate AI-based motion tracking directly into MRI scanner software for real-time correction
- Validate your algorithms using advanced MR physics setups and optical tracking (gold standard method)
- Collaborate with experts in MR physics, AI, and clinical imaging to ensure robust translation to 3T and 7T systems
- Present and publish your findings at top-tier machine learning/MRI journals and conferences
What you’ll learn (hands-on skills)
- Physics-informed neural networks for motion estimation and real-time control
- Simulation and processing of large-scale multi-coil k-space datasets
- High-performance GPU training, model optimization, and deployment via Siemens FIRE/ICE frameworks
- Sequence programming and real-time MRI system integration
- Scientific writing, experimental design, and interdisciplinary teamwork bridging AI, physics, and medicine
Your research environment
You will join the High-Field MR Center (HFMRC) at the Medical University of Vienna, an internationally recognized hub for ultra-high-field MRI method development. The center houses cutting-edge 3T and 7T MRI scanners, optical tracking system, and multi-coil shimming hardware, providing a unique environment to bring AI into the MRI scanner itself.
Key benefits:
- High-quality mentoring and being a part of the large team of many phds and postdocs
- Interdisciplinary collaboration with physicists, computer scientists, and clinicians
- Access to state-of-the-art hardware
- Career preparation for medical AI, imaging physics, or applied research
Required: Programming experience (Python, MATLAB), background in computer science, biomedical engineering, physics, or mathematics.
Advantageous: Experience with MRI data processing, deep learning frameworks (PyTorch, TensorFlow), and signal modelling.
Beneficial: Interest in medical imaging, high-performance computing, and real-time system integration.
- AI/ML
- Biomedical engineering
- Medical physics and/or Biophysics
This FWF-funded PhD project focuses on developing and deploying DL approaches for real-time B0-field stabilisation at 7T MR Scanner. The candidate will further developed our DL solution for prediction of changes of B0 field due to subject motion and deploy it directly at the scanner. The project integrates physics-based modelling and software engineering with AI to improve MR image and spectroscopic data quality at ultra-high field MRI systems.
What you’ll work on
- Develop deep neural networks to predict and correct dynamic B0-field changes caused by head motion and scanner drift
- Combine physics-based modelling with data-driven learning to control spherical harmonic and multi-coil shim systems
- Implement real-time field correction algorithms integrated with MRI sequences (EPI, MRSI)
- Design and test hybrid MR navigator strategies for frequency drift tracking
- Collaborate with engineers, AI researchers, and clinicians to translate your methods to clinical MRI scanners
- Present and publish your findings at top-tier machine learning/MRI journals and conferences
What you’ll learn (hands-on skills)
- Deep learning for field estimation, spatiotemporal modelling, and control systems
- Quantitative MR physics, B0 shimming principles, and magnetic field mapping
- Data simulation and augmentation for semi-synthetic MR field datasets
- Real-time integration and deployment of AI models within MRI scanner software
- Advanced scientific communication, hypothesis testing, and reproducible ML workflows
Your research environment
You will join the High-Field MR Center (HFMRC) at the Medical University of Vienna, an internationally recognized hub for ultra-high-field MRI method development. The center houses cutting-edge 3T and 7T MRI scanners, optical tracking system, and multi-coil shimming hardware, providing a unique environment to bring AI into the MRI scanner itself.
Key benefits:
- High-quality mentoring and being a part of the large team of many phds and postdocs
- Interdisciplinary collaboration with physicists, computer scientists, and clinicians
- Access to state-of-the-art hardware
- Career preparation for medical AI, imaging physics, or applied research
Programming experience (Python, MATLAB), background in computer science, physics, or related field.
Advantageous: Knowledge of MR physics, magnetic field modelling, or signal processing.
Beneficial: Familiarity with AI methods for image reconstruction or field estimation.

- Biochemistry
- Bioinformatics
- Molecular and cell biology
The pigmentation of the skin is an important regulator of interactions between the organism and the environment. The main pigment of the skin, melanin, is produced by cells known as melanocytes and subsequently transferred to epithelial cells. This mechanism of skin pigmentation is conserved in humans and other vertebrates. Previous studies of our group have suggested that another, yet uncharacterized mode of epithelial pigmentation is active in specialized skin structures of amphibians and possibly other vertebrates. The aim of the project is to determine the mechanism, function and evolution of non-canonical skin pigmentation. Recombinant protein expression, in vitro assays of enzymatic activities and investigations of samples from gene-edited Xenopus frogs will be used to determine the molecular control of non-canonical pigment formation. Furthermore, advanced methods of comparative genomics and molecular phylogenetics will be applied. This project will characterize a unique mode of epithelial pigmentation and may help to define novel functions of skin pigments in vertebrates.
What you’ll work on
- Investigate a novel, non-canonical mechanism of epithelial pigmentation in amphibian skin
- Express and characterize recombinant proteins involved in pigment formation
- Perform in vitro enzyme activity assays and analyze gene-edited Xenopus models
- Apply comparative genomics and molecular phylogenetics to study pigment evolution across vertebrates
- Integrate molecular, biochemical, and evolutionary data to uncover new functions of skin pigments
What you’ll learn (and get really good at)
- Designing, executing, and publishing original research projects
- Recombinant protein expression and biochemical activity assays
- Genome editing and functional analysis in Xenopus
- Comparative genomics, molecular phylogenetics, and data interpretation
- Scientific writing, presentation, and effective communication of results
Your research environment
You will join an internationally connected research group exploring the molecular and evolutionary biology of skin pigmentation. The project includes active collaborations with leading international partners, providing access to a wide range of expertise, model systems, and analytical technologies.
Master in molecular biology, biochemistry or related field
Advantage: Experience in molecular phylogenetics or biochemistry of enzymes

- AI/ML
- Animal research
- Bioinformatics
- Cancer
- Computer lab
- Human research
- Immunology
- Mathematics and/or Statistics
- Wet lab
Metastasis is the leading cause of cancer related deaths. Specific cancers have a prevalence to metastasize to specific organs. The underlying mechanisms for this organotropism are poorly understood. In this project you will investigate how tumor and metastatic cells and their immune-microenvironment are different in different host organs and how malignant cells remodel distant tissue niches. We are focusing on finding ways to empower the immune system to combat metastasis using cutting-edge single-cell and spatial omics technologies. We use clinically highly relevant disease models of metastasis and patient samples to investigate the complex interplays of tumor and immune cells in metastasis. The use of modern omics technologies such as single-cell RNA-seq and spatial transcriptomics and protein profiling makes it possible to discover new tumor immune cell interactions in metastasis that have not been described before. By studying these interactions between metastatic cancer cells and immune cells, we hope to uncover new insights that could lead to improved treatments for cancer patients.
We offer two PhD positions: one is more focused on understanding tumor-immune interactions using computational approaches and the other is using organoid systems to understand the impact of tumor heterogeneity on metastasis and metastatic niches.
What you´ll learn (and get really good at)
Our interdisciplinary approach combines different expertise to tackle the complex interplay of tumor and immune cells in metastasis including experimental and computational biology, as well as translational research. As a member of our team, you will receive training in both experimental and computational biology that are equally important skill sets for modern cancer researchers. You will be trained to generate and analyze single-cell and spatial omics data using the latest technologies, learn how to generate scientific hypothesis and to validate your hypothesis in appropriate advanced in vitro organoid and in vivo metastasis models systems. You will have the opportunity to present your work at conferences and collaborate with researchers from around the world, helping you grow professionally and make valuable connections in the field.
Your research enviroment
And as part of the unique SHIELD PhD program, you will have access to coordinated courses and workshops and a network of experts in immunology and cancer, providing even more opportunities for learning and collaboration.
Master in Biochemistry, Molecular biology, Immunology, Computational Biology or equivalent. Experience in computational biology and/or immunology are desirable, required excitement to work both computationally (analyzing single-omics and spatial omics data) and experimentally (working with mouse models), strong team spirit and high motivation to push the boundaries of metastatic research to improve patient outcomes.

- Animal research
- Endocrinology and Metabolism
- Human research
- Immunology
- Wet lab
The lung microenvironment is composed of structural and immune cells, that together shape the activity and plasticity of tissue resident macrophages. We and others discovered that immune responses within the lungs undergo drastic changes in age, with higher susceptibilities to prolonged inflammation following infection or the development of lung fibrosis. Together with an interdisciplinary team of scientists within the Excellence Cluster MetAGE (https://www.metage.at/en/) we attempt to test the idea that a loss of metabolic control in aging determines alterations of the immune response. Using a set of experimental model systems, we aim to investigate if nutritional interventions can reinstate metabolic control and “rejuvenate” immune homeostasis. Understanding the intricate interplay of metabolic control and the functionality of protective immunity will enable us to contribute to the concept of “healthy aging”.
What you’ll work on
- Map how aging and metabolism reprogram lung macrophages and immune homeostasis
- Test nutritional interventions in experimental models to restore metabolic control
- Profile immune responses using flow cytometry and single-cell RNA-seq
- Run macrophage functional assays and metabolic readouts
- Present results, co-author manuscripts, and help shape future studies
What you’ll learn (hands-on skills)
- Broad immunology toolkit: flow cytometry, scRNA-seq, macrophage assays
- Animal models of inflammation and infection (design, execution, analysis)
- Metabolic assays and data interpretation
- Reproducible analysis and clear scientific communication
Your research network
You will be part of the Excellence Cluster MetAGE (https://www.metage.at/en/), a collaboration of 25 research groups across Vienna and Graz with a vibrant student community, joint initiatives, and opportunities for cross-group interactions (e.g., seminars, workshops, short rotations).
Master’s degree (or equivalent) in biomedical sciences or related fields.
Advantage: Experience with mouse models, flow cytometry or immunological assays.

- AI/ML
- Bioinformatics
- Biomedical engineering
- Computer lab
- Data Science
Temporomandibular disorders (TMD) are a common cause of orofacial pain and often co-occur with headaches. They typically begin in early adulthood, impair core daily functions such as eating and speaking, and can lead to long, costly treatment journeys. TMDs occur more frequently in women, yet the underlying mechanisms remain unclear. Differences in TMJ morphology between women and men may drive functional and biomechanical differences. This FWF-funded project combines machine learning and physics-based simulation to uncover how morphology, muscle coordination, and joint loading interact to produce sex differences in mechanical stress and pain risk. We will develop machine learning–enhanced reduced-coordinate hybrid finite element – rigid body models of the TMJ for fast, personalized biomechanical simulation. Models will be trained and validated on multimodal data comprising MRI-based morphology, jaw-motion tracking, EMG, and bite-force recordings. In parallel, reinforcement learning will generate realistic muscle activations and motion patterns. Together, these approaches form digital twins of the TMJ to study sex-specific loading and predict individual risk factors for pain. Two fully funded PhD positions are available:
• PhD 1: Reduced-coordinate hybrid modeling and data-driven analysis linking TMJ morphology to mechanical stress.
• PhD 2: Reinforcement learning for motor control and simulation of muscle coordination in jaw function.
What you’ll work on
- Develop AI-based models for personalized simulation of dental biomechanics
- Apply deep learning to analyze multimodal data (MRI, EMG, motion tracking)
- Implement and test reinforcement learning algorithms for motor control and adaptive movement strategies
- Create explainable AI pipelines and calibrate hybrid models combining data-driven and physics-based methods
- Collaborate with engineers, clinicians, and data scientists to translate computational insights into clinical applications
What you’ll learn (hands-on skills)
- Machine and deep learning for biomechanical and imaging data
- Reduced-coordinate finite element modeling for personalized simulation
- Reinforcement learning for motor control and movement optimization
- Multimodal data integration and advanced signal processing
- Explainable AI, model interpretability, and rigorous model calibration
Your research environment
You will join the Competence Center Artificial Intelligence in Dentistry and the Comprehensive Center for AI in Medicine. The project involves collaboration with researchers at Clemson University (USA) and the University of British Columbia (Canada). We offer a vibrant, interdisciplinary setting at the interface of AI, engineering, and clinical dentistry, with unique patient datasets, high-performance computing, and ample opportunities to interact with clinical and ML/biomechanics experts.
• MSc (or equivalent) in Biomedical/Mechanical Engineering, Computer Science, Physics, or related field
• Strong programming and analytical skills (Python, PyTorch/TensorFlow, Java)
• Motivation to apply AI and simulation methods to human biomechanics
Advantage:
Experience with finite element modeling, reduced coordinate modeling, reinforcement learning, or medical imaging/biomechanical data.

- Animal research
- Immunology
- Molecular and cell biology
- Wet lab
Allergies are a global health problem that are driven by misdirected adaptive immune responses against mostly harmless proteins, derived for instance from food, pollen or insects. Mast cells are fascinating immune cells that are central for allergic reactions. They can be primed by allergen-specific antibodies and then immediately respond to allergen exposure by production of inflammatory mediators that cause symptoms like sneezing, itching or diarrhea. Even though mast cells have been known for over a century, many of their unique biological properties are still enigmatic.
In the proposed PhD projects, we will use in vitro primary mast cell models combined with a multi-omics strategy to systematically characterize and dissect transcriptional, epigenetic and metabolic programs that govern mast cell functions. This dataset will be the foundation of a new resource to substantially extend our understanding of mast cell biology. We will establish and apply cutting-edge tools, including CRISPR/Cas9 and shRNA-based gene perturbation, humanized mouse models and cell engineering to develop, test and validate innovative concepts for new treatments of allergies.
What you’ll work on
- Build and optimise primary mast cell models for mechanistic studies
- Run multi-omics experiments (transcriptomics, epigenomics, metabolomics) and integrate results
- Perform CRISPR/Cas9 and shRNA perturbations to map regulators of mast-cell function
- Contribute to humanized mouse and cell-engineering workflows for therapy concepts
- Present findings, co-author manuscripts, and help shape future project directions
What you’ll learn (hands-on skills)
- Primary immune cell culture and assay design
- Multi-omics data generation and interpretation
- Flow cytometry, ELISA, PCR, and standard molecular biology
- Practical genome editing (CRISPR/shRNA) and validation strategies
- Clear scientific storytelling (talks, posters, figures)
Your research environment
- International, interdisciplinary team focused on allergy and immune mechanisms, located at AKH
- Close collaboration with experts in immunology, genomics, and bioinformatics
We are looking for two PhD candidates for this project.
• Master’s degree (or equivalent) in biology, molecular biology, immunology, medicine or a related life science field.
• Experience in basic molecular biology techniques (e.g. ELISA, PCR, flow cytometry, etc)
• Experience in primary cell culture or/and animal work (mice) would be a plus
• Interest in immunology and enthusiasm about science

- Human research
- Medical physics and/or Biophysics
- Neuroscience
This PhD position will be a part of the clinical research group EPICONN, which will investigate the epileptogenic connectome with the overall goal to develop biomarkers to predict the best treatment options for people with epilepsy to forward personalised medicine. Our work packages mission aims are: We will combine quantitative 7T magnetic resonance spectroscopic imaging (MRSI) and FDG-positron emission tomography (PET) into a novel neurochemical imaging protocol for the description of metabolic changes in epileptogenic networks. We will generate novel metabolic insights about the rule of glutamate (Glu) and glutamine (Gln) in brain networks. Our metabolic findings will be correlated to functional connectivity findings (EEG, fMRI, resting state fMRI) and used for the deep learning analysis-based development of biomarkers. To this end, this prospective PhD position will implement and validate a new 7T MRSI study protocol that will be applied together with quantitative FDG-PET (another PhD project) to a cohort of 300 patients. Volume-of-interest and voxel-wise statistical analysis comparing MRSI and PET and metabolic connecome modelling will be the foundation for the development of machine-learning based biomarkers within the clinical research group. In summary, the expected work over the course of the PhD will be the acquisition and analysis of 7T MRSI data, its correlation to FDG-PET, and the integration of the metabolic connectome into the overall project.
What you’ll work on
- Implement and validate a 7T MRSI protocol and acquire/analyse patient data
- Integrate MRSI + FDG-PET and link metabolic findings with EEG/fMRI/rs-fMRI connectivity
- Build and evaluate ML/deep-learning biomarkers for personalised treatment decisions
- Collaborate closely with clinicians, imaging scientists, and data scientists; present at conferences
What you’ll learn (hands-on skills)
- 7 Tesla MRI operation, MRSI data processing
- Medical image analysis, connectomics, and scientific data presentation
- Multimodal data fusion and rigorous, reproducible research workflows
Your research network
This position is part of the 4-year clinical research group EPICONN that develops connectome biomarkers for epilepsy—working alongside 10 project staff, including six other PhD students. You’ll benefit from close collaboration, regular events, annual retreats, and dedicated training programmes for learning and networking.
- Master in biomedical engineering, medical informatics, physics, or any related subjects
- Basic knowledge of programming (esp. C++, Python, MATLAB, R) and statistics
- Organisation skills (scheduling, data management)
Advantage:
- Experience with medical imaging techniques (acquisition and processing)
- Experience with machine learning and/or connectomics
- Affinity for teamplay Previous experience in epilepsy research
- Animal research
- Biochemistry
- Cancer
- Drug targets and/or drug development
- Endocrinology and Metabolism
- Molecular and cell biology
- Wet lab
We are looking for two enthusiastic PhD students (m/f/d) to join our CRC-Res consortium funded by the Ludwig Boltzmann Gesellschaft. The consortium aims to elucidate how stromal and immune components of the tumor microenvironment drive therapy resistance in metastatic colorectal cancer, and to develop novel next-generation therapies to prevent and overcome drug resistance. These positions offer a unique opportunity to participate in preclinical and translational research within a multidisciplinary consortium including basic researchers, data scientists and clinicians offering the exclusive chance to translate our findings into early-phase clinical trials and to advance cancer therapy. Research will integrate patient-derived 3D tumor avatars and organoids with single cell and spatial OMICS, CRISPR/Cas9 genome engineering, proteomics and mouse models for preclinical validation.
The three laboratories offering the positions are part of the Center of Cancer Research (Dr. Sibilia, MedUni), the Department of General Surgery (Dr. Bergmann, MedUni, see seperate ad) and the Department of Biological Sciences and Pathobiology (Dr. Manieri, Vetmeduni).
What we offer
Multidisciplinary, international and collaborative consortium to develop research projects exploring drug resistance using cutting edge technologies in a supportive environment offering development opportunities in an inspiring, and international setting
Your Profile
o Master’s degree in a relevant biomedical subject
o Laboratory experience in basic molecular biology, biochemistry, cell culture techniques
- Animal research
- Biochemistry
- Cancer
- Drug targets and/or drug development
- Endocrinology and Metabolism
- Molecular and cell biology
- Wet lab
We are looking for three enthusiastic PhD students (m/f/d) to join our CRC-Res consortium funded by the Ludwig Boltzmann Gesellschaft. The consortium aims to elucidate how stromal and immune components of the tumor microenvironment drive therapy resistance in metastatic colorectal cancer, and to develop novel next-generation therapies to prevent and overcome drug resistance. These positions offer a unique opportunity to participate in preclinical and translational research within a multidisciplinary consortium including basic researchers, data scientists and clinicians offering the exclusive chance to translate our findings into early-phase clinical trials and to advance cancer therapy. Research will integrate patient-derived 3D tumor avatars and organoids with single cell and spatial OMICS, CRISPR/Cas9 genome engineering, proteomics and mouse models for preclinical validation.
The three laboratories offering the positions are part of the Center of Cancer Research (Dr. Sibilia, MedUni), the Department of General Surgery (Dr. Bergmann, MedUni) and the Department of Biological Sciences and Pathobiology (Dr. Manieri, Vetmeduni).
What we offer
Multidisciplinary, international and collaborative consortium to develop research projects exploring drug resistance using cutting edge technologies in a supportive environment offering development opportunities in an inspiring, and international setting
Your Profile
o Master’s degree in a relevant biomedical subject
o Laboratory experience in basic molecular biology, biochemistry, cell culture techniques

- AI/ML
- Biomedical engineering
- Computer lab
- Human research
Losing one’s natural voice through chronic hoarseness or laryngeal surgery profoundly impacts communication, identity, and quality of life. At the intersection of medicine and artificial intelligence, this project explores how AI-driven voice conversion can help people speak again, as well as restore clarity, expressiveness, and individuality of pathological speakers.
The PhD candidate will contribute to the development of novel deep-learning methods for transforming disordered voices into natural-sounding, intelligible speech while preserving speaker identity. Using real clinical data from patients after laryngectomy and individuals with chronic dysphonia, the work will bridge speech signal processing, representation learning, and digital health innovation.
The research combines methodological AI development with impactful application, embedded in a collaboration between the Speech and Hearing Science Lab (SHS Lab, Medical University of Vienna) and the Signal Processing and Speech Communication Lab (SPSC, Graz University of Technology). Candidates will work with interdisciplinary teams in AI in medicine, access state-of-the-art compute infrastructure, and engage with international partners in speech technology and rehabilitation science.
This position offers the opportunity to shape a next-generation assistive technology, i.e., a digital speaking aid designed to enhance communication and social participation for people affected by speech pathologies. Join us in giving these people back their voices.
What you’ll work on
- Develop deep-learning voice conversion methods for pathological-to-natural speech
- Advance speech signal processing for recognition, enhancement (incl. audio-video), synthesis, and representation learning
- Design and execute experiments on HPC/GPU infrastructure
- Build evaluation pipelines (objective metrics, perceptual tests, human-in-the-loop)
- Collaborate with clinicians, linguists, and engineers; present results and co-author papers
What you’ll learn (hands-on skills)
- Advanced speech processing & generative modelling (E.g., automated speech recognition, speech enhancement, text-to-speech, voice conversion)
- Deep Learning engineering (E.g., with PyTorch), data curation, experiment design on HPC
- Perceptual testing & benchmarking
- Interdisciplinary collaboration across medicine, linguistics, and computer science
Your research network
This position spans the SPSC Lab (Graz) and the SHS Lab/CAIM (Vienna) within a coordinated program across partner groups. You will work alongside other PhD students in speech, AI, and clinical research in this cross-lab project, share access to HPC/GPU infrastructure, and a steady rhythm of group meetings that build skills and visibility across the network. International ties with Nagoya University (Japan) and the Unite! Graduate School add even more options for exchange and short research rotations, mentoring, and cross-institutional coursework.
• M.Sc. degree in a relevant field (Electrical Engineering, Computer Science, Information & Computer Engineering, Electrical Engineering & Audio Engineering).
• Experience in speech signal processing, preferably in voice conversion.
• Willingness and eligibility to work in Graz as well as in Vienna, Austria.
• Willingness and eligibility to travel to internationally for scientific meetings.

- Animal research
- Cancer
- Immunology
- Molecular and cell biology
Lung adenocarcinoma (LUAD) is the most common form of lung cancer and remains the leading cause of cancer-related death worldwide. Despite advances in targeted therapies and immune checkpoint blockade, many LUADs remain resistant to treatment, underscoring the urgent need for new therapeutic strategies.
Our lab has discovered that partial loss of the inflammatory regulator A20 (TNFAIP3) in mice profoundly alters the lung tumor microenvironment: it promotes the formation of tertiary lymphoid structures (TLS), increases B cell infiltration, and drives local antibody production in KRAS-driven lung tumors. These findings point to an overlooked contribution of B cells and TLS in shaping anti-tumor immunity.
This PhD project will use advanced mouse models, single-cell and spatial profiling, and functional perturbation of B cells to uncover how B cells and TLS influence lung tumor progression and responses to immunotherapy. By clarifying these mechanisms, the project aims to establish whether targeting A20-regulated pathways and harnessing B cell–TLS biology can be developed into new immunotherapeutic strategies to overcome resistance in LUAD.
What you’ll work on
- Establish and analyse mouse models of KRAS-driven lung cancer to study B cell–driven immune responses
- Use depletion/adoptive transfer experiments to define B-cell impact on tumor progression
- Apply scRNA-seq and, as feasible, spatial analyses to dissect TLS biology and B-cell function
- Mine and integrate data from TCGA, GEO, and internal datasets
- Employ standard molecular & cellular biology methods
- Present results, contribute to manuscripts, and help shape future grant proposals
What you’ll learn (hands-on skills)
- In-vivo tumor immunology using state-of-the-art models
- Single-cell and spatial profiling/analysis of immune microenvironments
- Data integration across public and in-house datasets
- Clear scientific communication (talks, posters, figures, papers)
Your research network
This position sits within MedUni Vienna’s tumor-immunology network and a 4-year programme aligned with partnering labs. You’ll work alongside a cohort of PhD students and postdocs across collaborating groups, with close cross-lab projects, shared access to core facilities (imaging, genomics, flow cytometry, animal hubs), and a steady rhythm of joint seminars, methods clubs, retreats, and training programmes that foster learning, feedback, and visibility across the network.
- Master’s degree in life sciences (immunology, molecular biology, biotechnology, or related fields)
- Strong interest in tumor immunology
Advantage:
- Experience in molecular/cell biology techniques; mouse handling
- Basic programming skills (R or Python) and interest in data analysis